Dividing Clusters (aka compartmental cells)
Related: Compartmentalized Cells, ContactInternal Plugin, and related XML
In compartmental models, a single cell is actually composed of several compartments.
Each compartment is somewhat like an individual cell in that its behaviors
can be specified independently, yet all the compartments can be treated as a single entity called a “cluster.”
A cluster is a collection of compartment cells with the same clusterId. If you use “simple” (non-compartmentalized) cells, then you can check that each such cell has a distinct id and clusterId.
An example simulation can be found in CompuCellPythonTutorial/clusterMitosis.
Let’s look at how we can divide “compact,” that is, “blob-shaped,” clusters.
from cc3d.core.PySteppables import *
class MitosisSteppableClusters(MitosisSteppableClustersBase):
def __init__(self, frequency=1):
MitosisSteppableClustersBase.__init__(self, frequency)
def step(self, mcs):
for cell in self.cell_list:
cluster_cell_list = self.get_cluster_cells(cell.clusterId)
print("DISPLAYING CELL IDS OF CLUSTER ", cell.clusterId, "CELL. ID=", cell.id)
for cell_local in cluster_cell_list:
print("CLUSTER CELL ID=", cell_local.id, " type=", cell_local.type)
mitosis_cluster_id_list = []
for compartment_list in self.clusterList:
# print( "cluster has size=",compartment_list.size())
cluster_id = 0
cluster_volume = 0
for cell in CompartmentList(compartment_list):
cluster_volume += cell.volume
cluster_id = cell.clusterId
# condition under which cluster mitosis takes place
if cluster_volume > 250:
# instead of doing mitosis right away we store ids for clusters which should be divide.
# This avoids modifying cluster list while we iterate through it
mitosis_cluster_id_list.append(cluster_id)
for cluster_id in mitosis_cluster_id_list:
self.divide_cluster_random_orientation(cluster_id)
# # other valid options - to change mitosis mode leave one of the below lines uncommented
# self.divide_cluster_orientation_vector_based(cluster_id, 1, 0, 0)
# self.divide_cluster_along_major_axis(cluster_id)
# self.divide_cluster_along_minor_axis(cluster_id)
def update_attributes(self):
# compartments in the parent and child clusters are
# listed in the same order so attribute changes require simple iteration through compartment list
compartment_list_parent = self.get_cluster_cells(self.parent_cell.clusterId)
for i in range(len(compartment_list_parent)):
compartment_list_parent[i].targetVolume /= 2.0
self.clone_parent_cluster_2_child_cluster()
The steppable is quite similar to the mitosis steppable which works for
non-compartmental cells. This time, after mitosis happens, you
have to reassign properties of children compartments and of parent
compartments which usually means iterating over a list of compartments.
Conveniently, this iteration is quite simple, and SteppableBasePy class
has a convenience function called get_cluster_cells which returns a list of cells
belonging to a cluster with a given clusterId:
compartment_list_parent = self.get_cluster_cells(self.parent_cell.clusterId)
The call above returns a list of cells in a cluster with clusterId
specified by self.parent_cell.clusterId. In the subsequent for-loop, we
iterate over a list of cells in the parent cluster and assign appropriate
values of volume constraint parameters. Notice that
compartment_list_parent is indexable (ie. we can access directly any
element of the list provided our index is not out of bounds).
for i in range(len(compartment_list_parent)):
compartment_list_parent[i].targetVolume /= 2.0
Notice that nowhere in the update attribute function we have modified cell types. This is because, by default, the cluster mitosis module assigns cell types to all the cells of the child cluster and it does it in such a way that the child cell looks like a quasi-clone of the parent cell.
The next call in the update_attributes function is
self.clone_parent_cluster_2_child_cluster(). This copies all the attributes
of the cells in the parent cluster to the corresponding cells in the
child cluster. If you would like to copy attributes from parent to child
cell skipping select ones you may use the following code:
compartment_list_parent = self.get_cluster_cells(self.parent_cell.clusterId)
compartment_lis_child = self.get_cluster_cells(self.child_cell.clusterId)
self.clone_cluster_attributes(source_cell_cluster=compartment_list_parent,
target_cell_cluster=compartment_list_child,
no_clone_key_dict_list=['ATTR_NAME_1', 'ATTR_NAME_2'])
where clone_cluster_attributes function allows specification of this
attributes are not to be copied (in our case cell.dict members
ATTR_NAME_1 and ATTR_NAME_2 will not be copied).
Finally, if you prefer manually setting the parent and child cells, you would use the following code:
class MitosisSteppableClusters(MitosisSteppableClustersBase):
def __init__(self, frequency=1):
MitosisSteppableClustersBase.__init__(self, frequency)
def step(self, mcs):
for cell in self.cell_list:
cluster_cell_list = self.get_cluster_cells(cell.clusterId)
print("DISPLAYING CELL IDS OF CLUSTER ", cell.clusterId, "CELL. ID=", cell.id)
for cell_local in cluster_cell_list:
print("CLUSTER CELL ID=", cell_local.id, " type=", cell_local.type)
mitosis_cluster_id_list = []
for compartment_list in self.clusterList:
# print( "cluster has size=",compartment_list.size())
cluster_id = 0
cluster_volume = 0
for cell in CompartmentList(compartment_list):
cluster_volume += cell.volume
cluster_id = cell.clusterId
# condition under which cluster mitosis takes place
if cluster_volume > 250:
# instead of doing mitosis right away we store ids for clusters which should be divide.
# This avoids modifying cluster list while we iterate through it
mitosis_cluster_id_list.append(cluster_id)
for cluster_id in mitosis_cluster_id_list:
self.divide_cluster_random_orientation(cluster_id)
# # other valid options - to change mitosis mode leave one of the below lines uncommented
# self.divide_cluster_orientation_vector_based(cluster_id, 1, 0, 0)
# self.divide_cluster_along_major_axis(cluster_id)
# self.divide_cluster_along_minor_axis(cluster_id)
def updateAttributes(self):
parent_cell = self.mitosisSteppable.parentCell
child_cell = self.mitosisSteppable.childCell
compartment_list_child = self.get_cluster_cells(child_ell.clusterId)
compartment_list_parent = self.get_cluster_cells(parent_cell.clusterId)
for i in range(len(compartment_list_child)):
compartment_list_parent[i].targetVolume /= 2.0
compartment_list_child[i].targetVolume = compartment_list_parent[i].targetVolume
compartment_list_child[i].lambdaVolume = compartment_list_parent[i].lambdaVolume
A Python helper for mitosis is available from Twedit++’s code snippets:
CC3D Python->Mitosis.
How It Works
While dividing non-clustered cells is straightforward, doing the same for clustered cells is more challenging. To divide non-clustered cells using the directional mitosis algorithm, we construct a line or a plane passing through the center of mass of a cell and pixels of the cell on one side of the line/plane end up in the child cell and the rest stays in the parent cell. Be sure to use the PixelTracker plugin with mitosis to enable this pixel manipulation. The orientation of the line/plane can be either specified by the user, or we can use CC3D’s built-in feature to calculate the orientation of the principal axes and divide either along the minor or major axis.
With compartmental cells, things get more complicated because: 1) Compartmental cells are composed of many subcells. 2) There can be different topologies of clusters. Some clusters may look “snake-like” and some might be compactly packed blobs of subcells. The algorithm which we implemented in CC3D works in the following way:
We first construct a set of pixels containing every pixel belonging to a cluster cell. You may think of it as a single “regular” cell.
We store volumes of compartments so that we know how big compartments should be after mitosis (they will be half of the original volume)
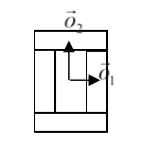
We calculate the center of mass of the entire cluster and calculate the vector offsets between the center of mass of a cluster and the center of mass of particular compartments as in the figure below:
Figure 7. Vectors \(\vec{o}_1\) and \(\vec{o}_2\) show offsets between center of mass of a cluster and center of mass particular compartments.
4) We pick a division line/plane for parent and child cells with offsets between cluster center of mass (after mitosis) and center of masses of clusters. We do it according to the formula:
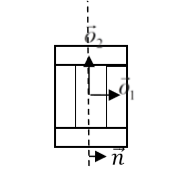
where \(\vec{p}\) denotes offset after mitosis from the center of mass of child (parent) clusters, \(\vec{o}\) is the orientation vector before mitosis (see picture above), and \(\vec{n}\) is a normalized vector perpendicular to division line/plane. If we try to divide the cluster along a dashed line as in the picture below:
Figure 8. Division of cell along dashed line. Notice the orientation of \(\vec{n}\) . The offsets after the mitosis for child and parent cell will be \(\vec{p}_1=\frac{1}{2}\vec{o}_1\) and \(\vec{p}_2=\vec{o}_2\) as expected because both parent and child cells will retain their heights, but these cells will also become twice as narrow.
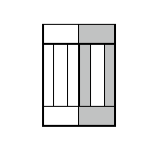
Figure 9.Child and parent cells after mitosis. The parent cell is the one with gray outer compartments.
The formula given above is heuristic. It gives a fairly simple way of assigning pixels of child/parent clusters to cellular compartments. It is not perfect, but the idea is to get the approximate shape of the cell after the mitosis, and, as the simulation runs, the cell shape will readjust based on constraints such as adhesion of focal point plasticity. Before continuing with mitosis, we check if the center of masses of the compartments belong to child/parent clusters. If the center of masses are outside their target pixels, we abandon mitosis and wait for readjustment of the cell shape, at which point the mitosis algorithm will pass this sanity check. For certain “exotic” shapes of cluster shapes, this mitosis algorithm may not work well or at all. In this case, we would have to write a specialized mitosis algorithm.
5) We divide clusters and knowing offsets from the child/parent cluster center of mass we assign pixels to particular compartments. The assignment is based on the distance of the particular pixel to the center of masses of clusters. The pixel is assigned to the compartment if its distance to the center of mass of the compartment is the smallest as compared to distances between centroids of other compartments. If the given compartment has reached its target volume and other compartments are underpopulated, we would assign pixels to other compartments based on the closest distance criterion. Although this method may result in some deviation from perfect 50-50 division of compartment volume, the cells will typically readjust their volume after a few MCS.
Figure 10. CC3D example of compartmental cell division. See also Demos/CompuCellPythonTutorial/clusterMitosis.